Guanzhong Gong
Unsupervised Anomaly Detection in Brain MRI via Disentangled Anatomy Learning
Dec 26, 2025Abstract:Detection of various lesions in brain MRI is clinically critical, but challenging due to the diversity of lesions and variability in imaging conditions. Current unsupervised learning methods detect anomalies mainly through reconstructing abnormal images into pseudo-healthy images (PHIs) by normal samples learning and then analyzing differences between images. However, these unsupervised models face two significant limitations: restricted generalizability to multi-modality and multi-center MRIs due to their reliance on the specific imaging information in normal training data, and constrained performance due to abnormal residuals propagated from input images to reconstructed PHIs. To address these limitations, two novel modules are proposed, forming a new PHI reconstruction framework. Firstly, the disentangled representation module is proposed to improve generalizability by decoupling brain MRI into imaging information and essential imaging-invariant anatomical images, ensuring that the reconstruction focuses on the anatomy. Specifically, brain anatomical priors and a differentiable one-hot encoding operator are introduced to constrain the disentanglement results and enhance the disentanglement stability. Secondly, the edge-to-image restoration module is designed to reconstruct high-quality PHIs by restoring the anatomical representation from the high-frequency edge information of anatomical images, and then recoupling the disentangled imaging information. This module not only suppresses abnormal residuals in PHI by reducing abnormal pixels input through edge-only input, but also effectively reconstructs normal regions using the preserved structural details in the edges. Evaluated on nine public datasets (4,443 patients' MRIs from multiple centers), our method outperforms 17 SOTA methods, achieving absolute improvements of +18.32% in AP and +13.64% in DSC.
* Accepted by Medical Image Analysis (2025)
COVID-MTL: Multitask Learning with Shift3D and Random-weighted Loss for Automated Diagnosis and Severity Assessment of COVID-19
Dec 31, 2020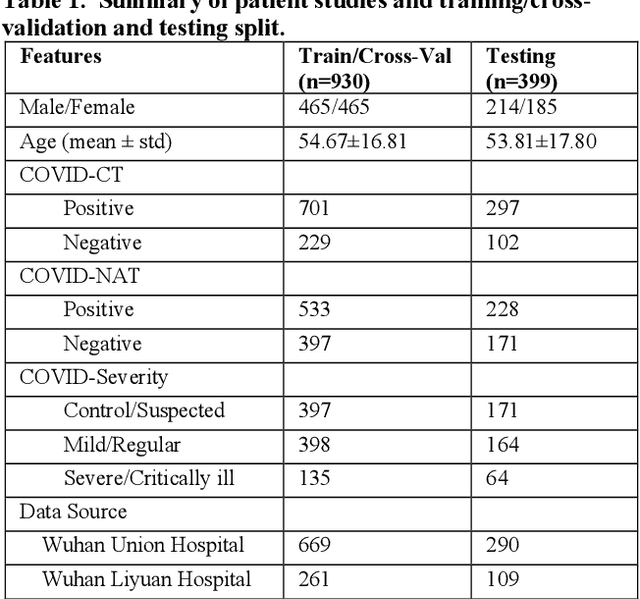
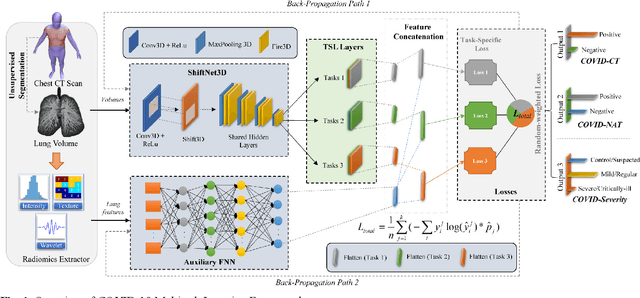
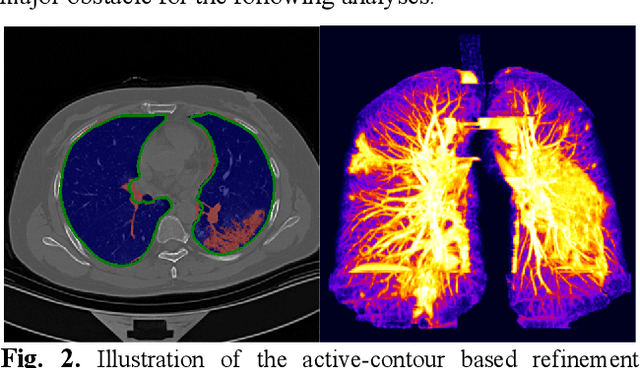
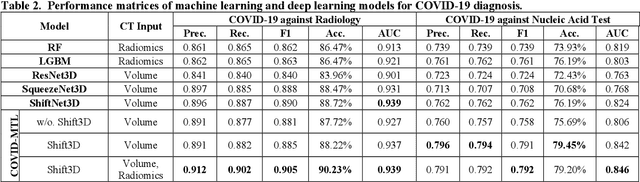
Abstract:There is an urgent need for automated methods to assist accurate and effective assessment of COVID-19. Radiology and nucleic acid test (NAT) are complementary COVID-19 diagnosis methods. In this paper, we present an end-to-end multitask learning (MTL) framework (COVID-MTL) that is capable of automated and simultaneous detection (against both radiology and NAT) and severity assessment of COVID-19. COVID-MTL learns different COVID-19 tasks in parallel through our novel random-weighted loss function, which assigns learning weights under Dirichlet distribution to prevent task dominance; our new 3D real-time augmentation algorithm (Shift3D) introduces space variances for 3D CNN components by shifting low-level feature representations of volumetric inputs in three dimensions; thereby, the MTL framework is able to accelerate convergence and improve joint learning performance compared to single-task models. By only using chest CT scans, COVID-MTL was trained on 930 CT scans and tested on separate 399 cases. COVID-MTL achieved AUCs of 0.939 and 0.846, and accuracies of 90.23% and 79.20% for detection of COVID-19 against radiology and NAT, respectively, which outperformed the state-of-the-art models. Meanwhile, COVID-MTL yielded AUC of 0.800 $\pm$ 0.020 and 0.813 $\pm$ 0.021 (with transfer learning) for classifying control/suspected, mild/regular, and severe/critically-ill cases. To decipher the recognition mechanism, we also identified high-throughput lung features that were significantly related (P < 0.001) to the positivity and severity of COVID-19.
Shape-Aware Organ Segmentation by Predicting Signed Distance Maps
Dec 09, 2019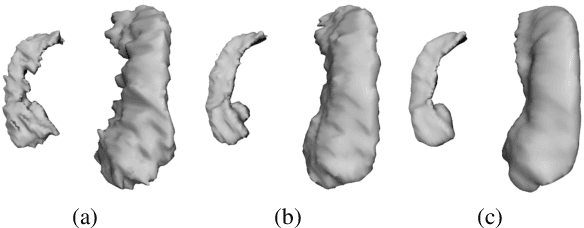
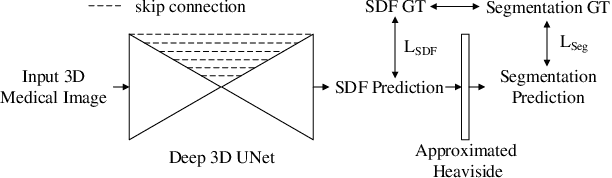
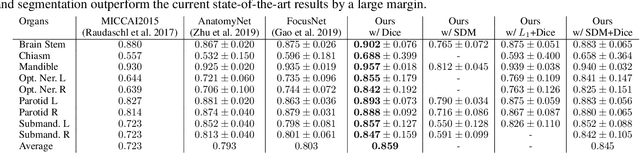
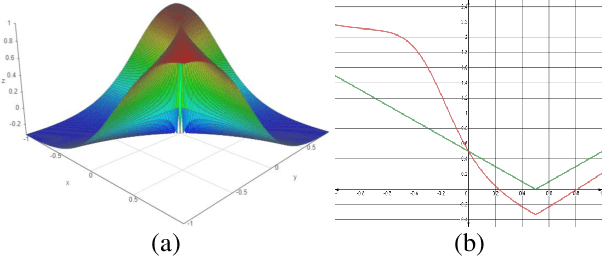
Abstract:In this work, we propose to resolve the issue existing in current deep learning based organ segmentation systems that they often produce results that do not capture the overall shape of the target organ and often lack smoothness. Since there is a rigorous mapping between the Signed Distance Map (SDM) calculated from object boundary contours and the binary segmentation map, we exploit the feasibility of learning the SDM directly from medical scans. By converting the segmentation task into predicting an SDM, we show that our proposed method retains superior segmentation performance and has better smoothness and continuity in shape. To leverage the complementary information in traditional segmentation training, we introduce an approximated Heaviside function to train the model by predicting SDMs and segmentation maps simultaneously. We validate our proposed models by conducting extensive experiments on a hippocampus segmentation dataset and the public MICCAI 2015 Head and Neck Auto Segmentation Challenge dataset with multiple organs. While our carefully designed backbone 3D segmentation network improves the Dice coefficient by more than 5% compared to current state-of-the-arts, the proposed model with SDM learning produces smoother segmentation results with smaller Hausdorff distance and average surface distance, thus proving the effectiveness of our method.
MMFNet: A Multi-modality MRI Fusion Network for Segmentation of Nasopharyngeal Carcinoma
Jan 22, 2019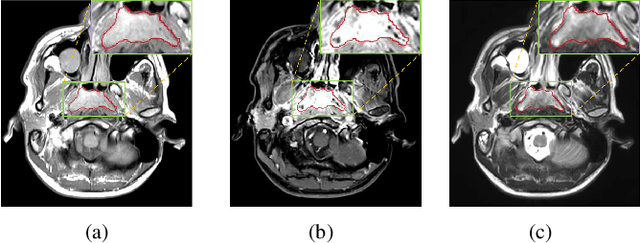
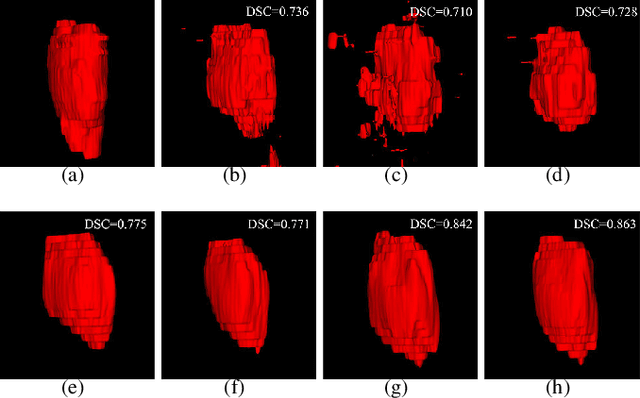
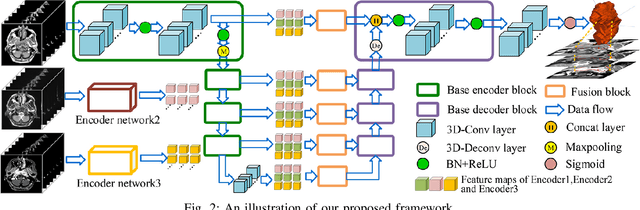
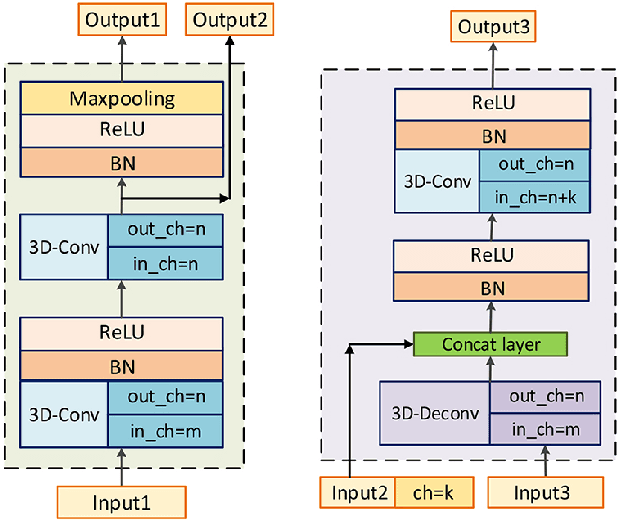
Abstract:Segmentation of nasopharyngeal carcinoma (NPC) from Magnetic Resonance Images (MRI) is a crucial step in NPC radiotherapy. However, manually segmenting of NPC is a time-consuming and labor-intensive task. Additionally, single-modality MRI generally cannot provide enough information for the accurate delineation of NPC. Therefore, a multi-modality MRI fusion network (MMFNet) based on three modalities of MRI (T1, T2 and contrast-enhanced T1) is proposed to complete accurate segmentation of NPC. In the MMFNet, the backbone is designed as a multi-encoder-based network, consisting of several modality-specific encoders and one single decoder. It can be used to well learn both low-level and high-level features used implicitly for NPC segmentation in each modality of MRI. A fusion block is proposed in the MMFNet to effectively fuse low-level features from multi-modality MRI. It firstly recalibrates features captured from multi-modality MRI, which will highlight informative features and regions of interest. Then, a residual fusion block is utilized to fuse weighted features before merging them with features from decoder to keep balance between high-level and low-level features. Moreover, a training strategy named self-transfer is proposed to initialize encoders for multi-encoder-based network. It can stimulate encoders to make full mining of modality-specific MRI. The proposed method can effectively make use of information in multi-modality MRI. Its effectiveness and advantages are validated by many experiments and comparisons with the related methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge